Non-coding Gene Mutations and the Relationship to the Spectrum of Neurodevelopmental Disorders
At Triple V, we love highlighting the research, writing, and work of our Fellows. This piece features a research paper by Fellow Natasha Russ-Stucki and reflects her growing work and interest in the field of neurodevelopmental psychology.
Russ1, T. R. Chinthapalli2, C. Zhu3, B. Ramirez4, N. Sun5 , A. Hassan61Arizona State University, Tempe, 2Apex High School, Apex, 3The Bear Creek School, Bellevue, 4University of Arkansas, Fayetteville, 5Northwood High School, Irvine, 6Aurora High School, Aurora, United States
Objectives: To identify the key non-coding genetic mutations connected to neurodevelopmental disorders, evaluate how mutations like RNU2-2 could alter gene regulation, and determine how these disruptions contribute together to ASD and related conditions.
Background: Neurodevelopmental disorders such as ASD, ADHD, Tourette’s Syndrome, and limited intellectual capacities show that there is strong genetic influence, however, many cases cannot be explained by protein-coding mutations alone, as a major portion of the risk lies in non-coding regulatory regions. Recent studies on whole-genome analyses have found that non-coding variants, including enhancer and promoter disruptions, intronic mutations, and microRNA seed-region changers, alter gene expression during fetal brain development by disrupting genes that are key to neurodevelopment. Non-coding genes alter many essential pathways for brain development. Together, the evidence shows that regulatory mutations are crucial to understanding neurodevelopmental diseases beyond protein-coding alterations.
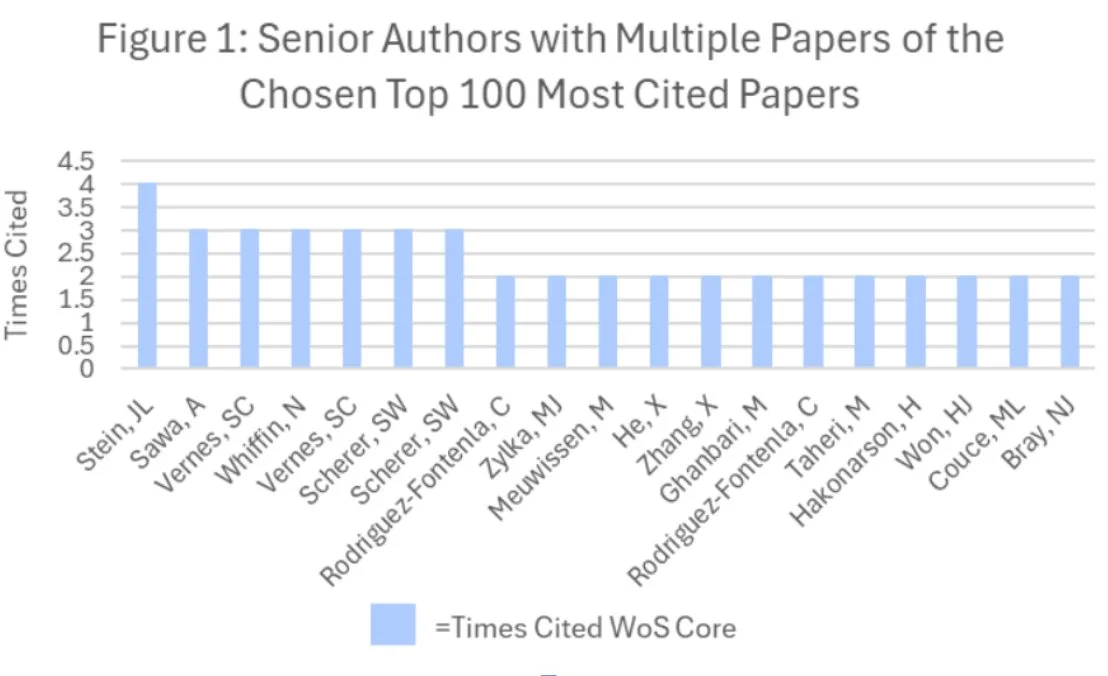
Figure 2:
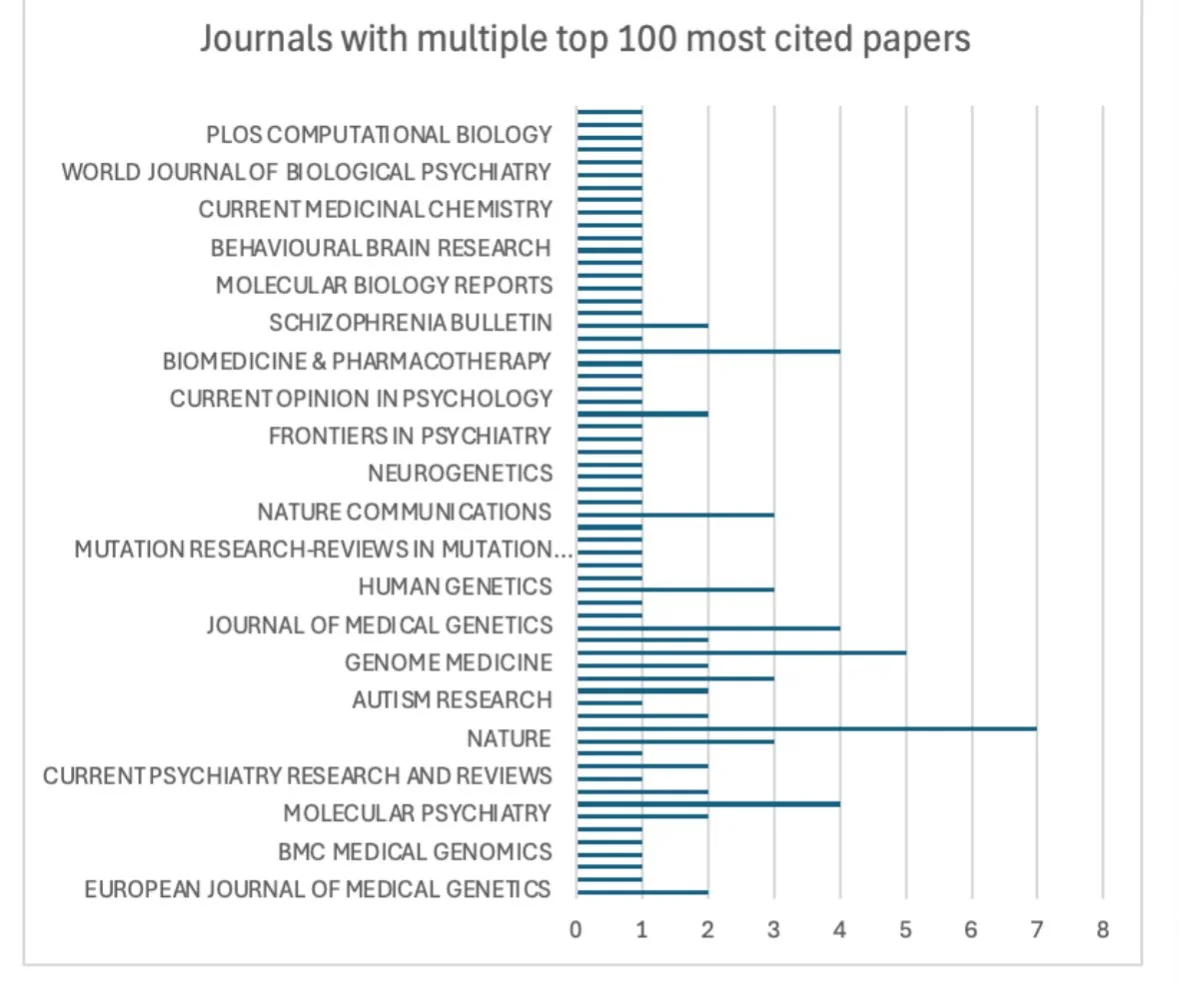
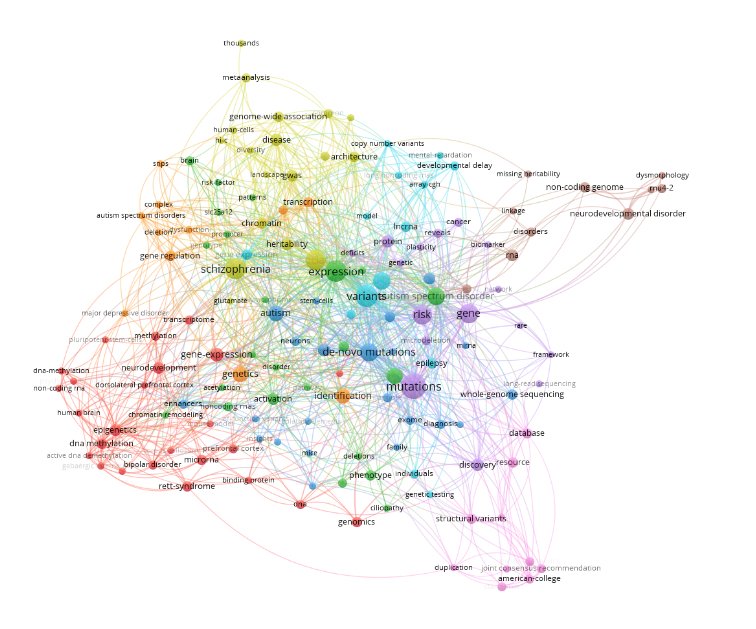
Methods: Using the R programming language (Version 4.5.2) in RStudio (Version 2025.09.2+418), we cleaned and analyzed bibliographic data of the top 100 articles with keywords collected from Web of Science. Low-information keywords were filtered out and all terminology standardized. We produced a keyword-trend plot, co-authorship, and co-citation networks in RStudio; senior author, institution, journal distribution, and method figures from extracted tables in Microsoft Excel; and keyword co-occurrence map in VOSviewer (Version 1.6.19).
Figure 3:
Results: There is a link between non-coding variants that significantly contribute to ASD and neurodevelopmental disorder risk. There is evidence to suggest that non-coding mutations alter gene regulation during fetal brain development by disrupting core neurodevelopmental pathways. The analysis showed a trend in specific non-coding RNAs, like snRNA, miRNAs, and antisense RNAs contributing to pathology. Overall non-coding genetic variants contribute to neurodevelopmental disorders by disrupting gene regulation, RNA processing, synaptic development, and neurogenesis.
Conclusions: The study concluded that small genetic changes can work together to raise autism risk specifically but their effect is small. Significant genes like MAPK3 and PTEN are associated with deletion syndrome/intellectual disability and developmental delay. More research is needed to identify which DNA changes are the most significant. Ultimately, more research is needed to develop these areas of interest, in order to uncover decisively what the impact of non-coding gene mutations on the the spectrum of neurodevelopmental disorders.